Software
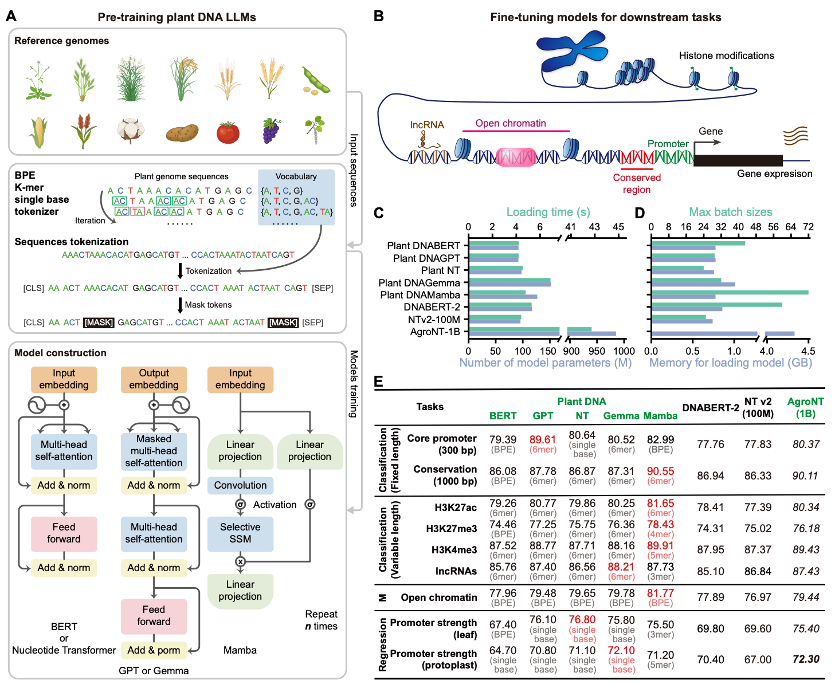
Plant DNA LLMs
Introduction
Chorus2
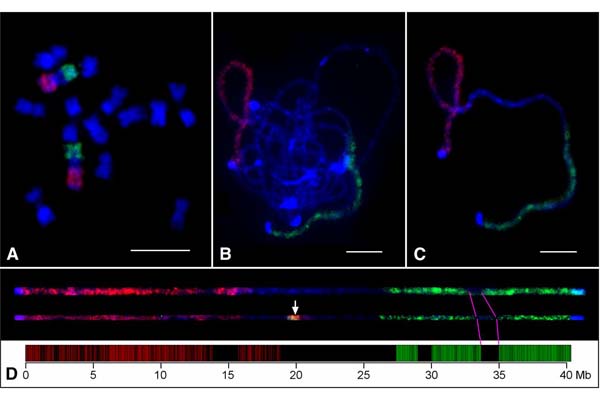
Design of genome-scale oligonucleotide-based probes for fluorescence in situ hybridization (FISH)
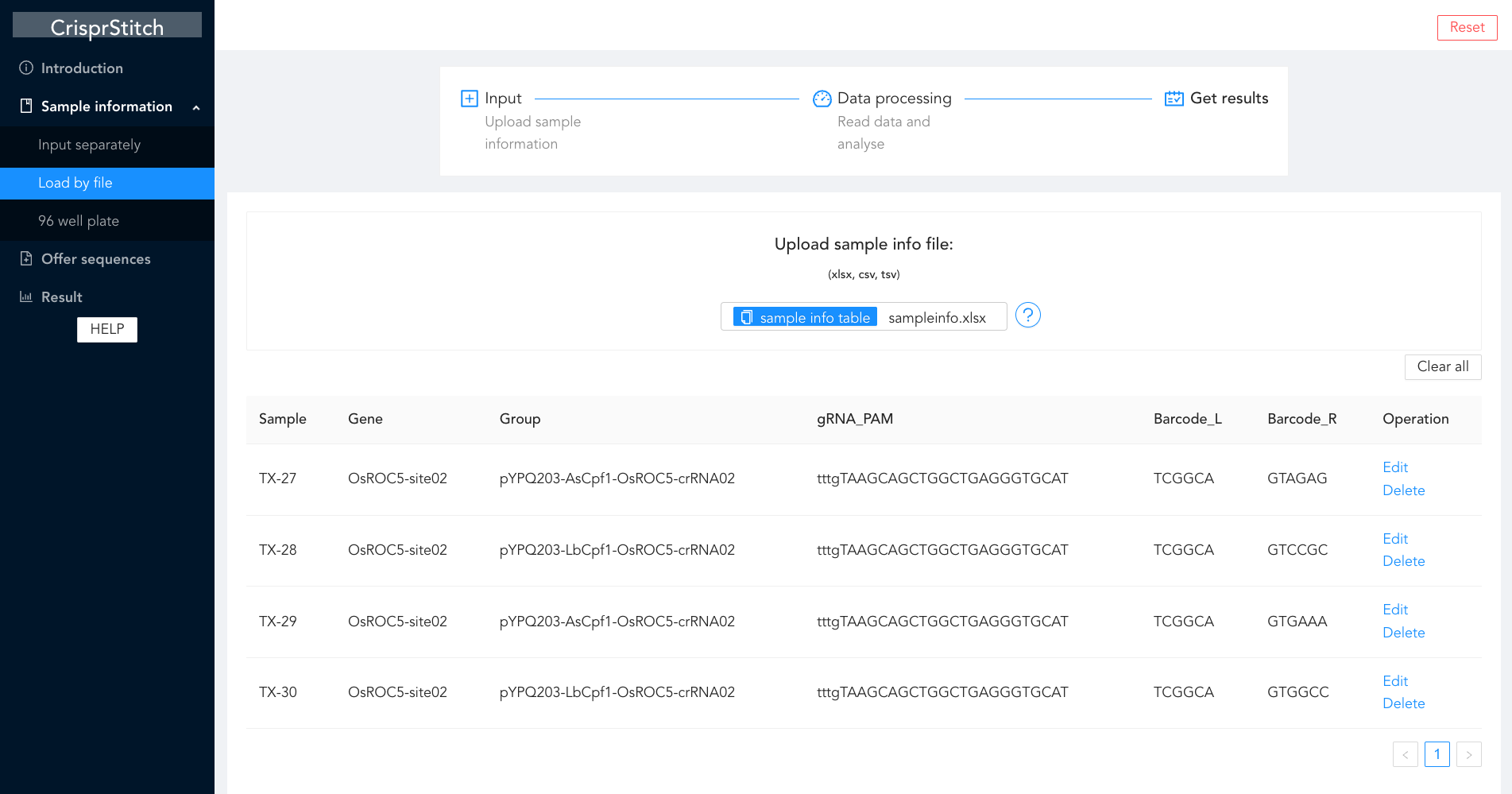
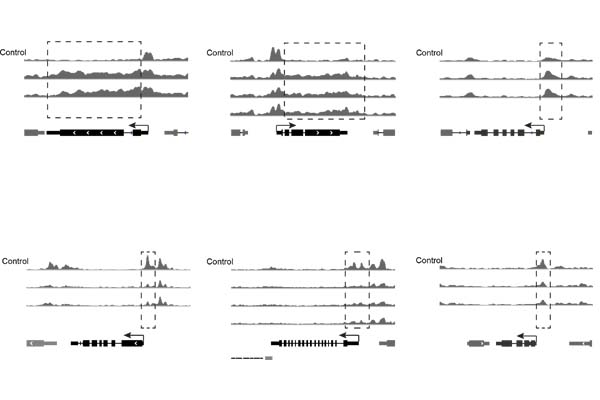
CrisprStitch
An integrated tool to fast evaluate the efficiency of CRISPR editing systems.
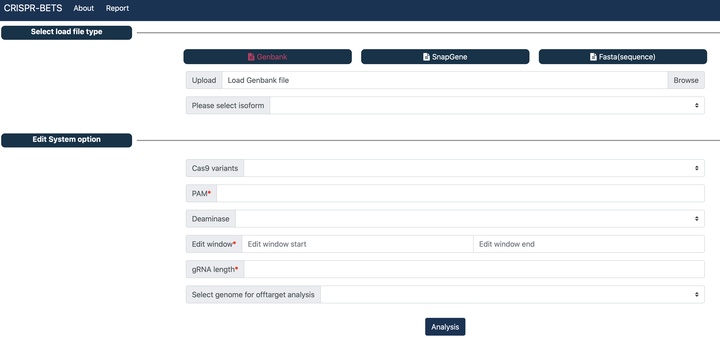
CRISPR-BETS
A gRNA design software for base editing knockout using stop codon.
Popera
A software specially designed for DHS identification. Github Introduction DNase I hypersensitive sites (DHSs) are regions of chromatin that are sensitive to cleavage by the...
Latest
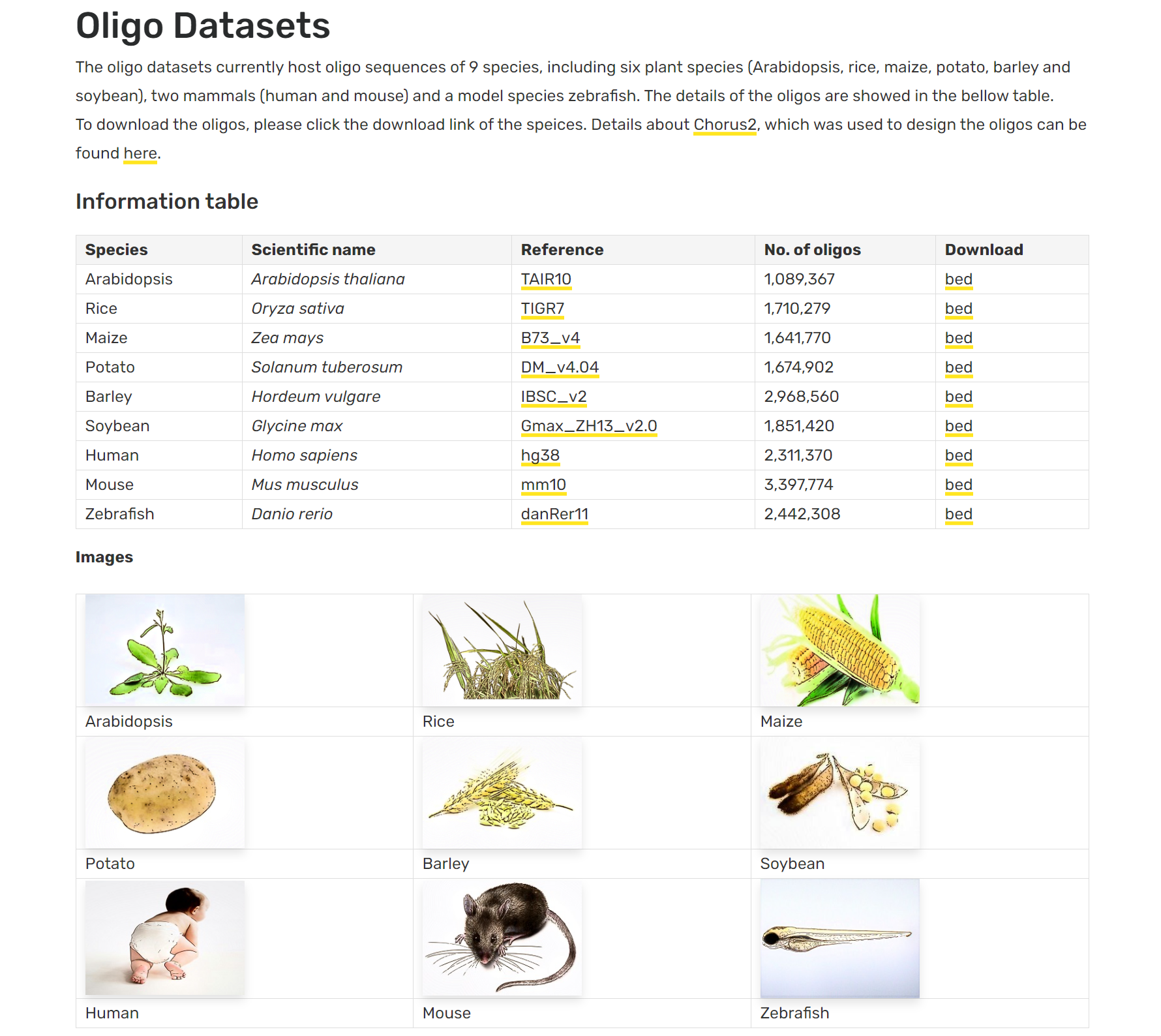
Chorus2 oligo datasets
Oligo datasets which host oligo sequences for Oligo-FISH. Details Introduction The oligo datasets currently host oligo sequences of nine species, including Arabidopsis, rice, maize, potato, barley, soybean, human, mouse and...
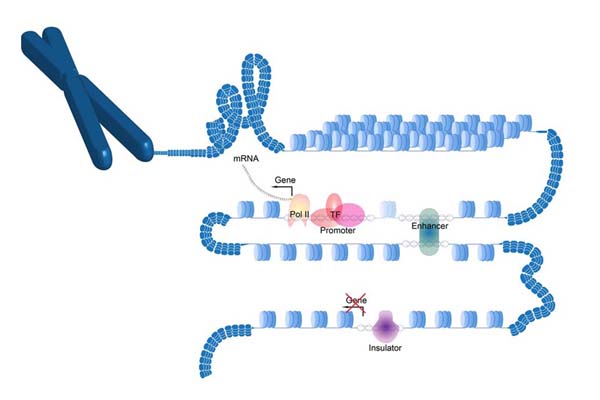
PlantDHS
PlantDHS: a database for DNase I hypersensitive sites (DHSs) in plants.
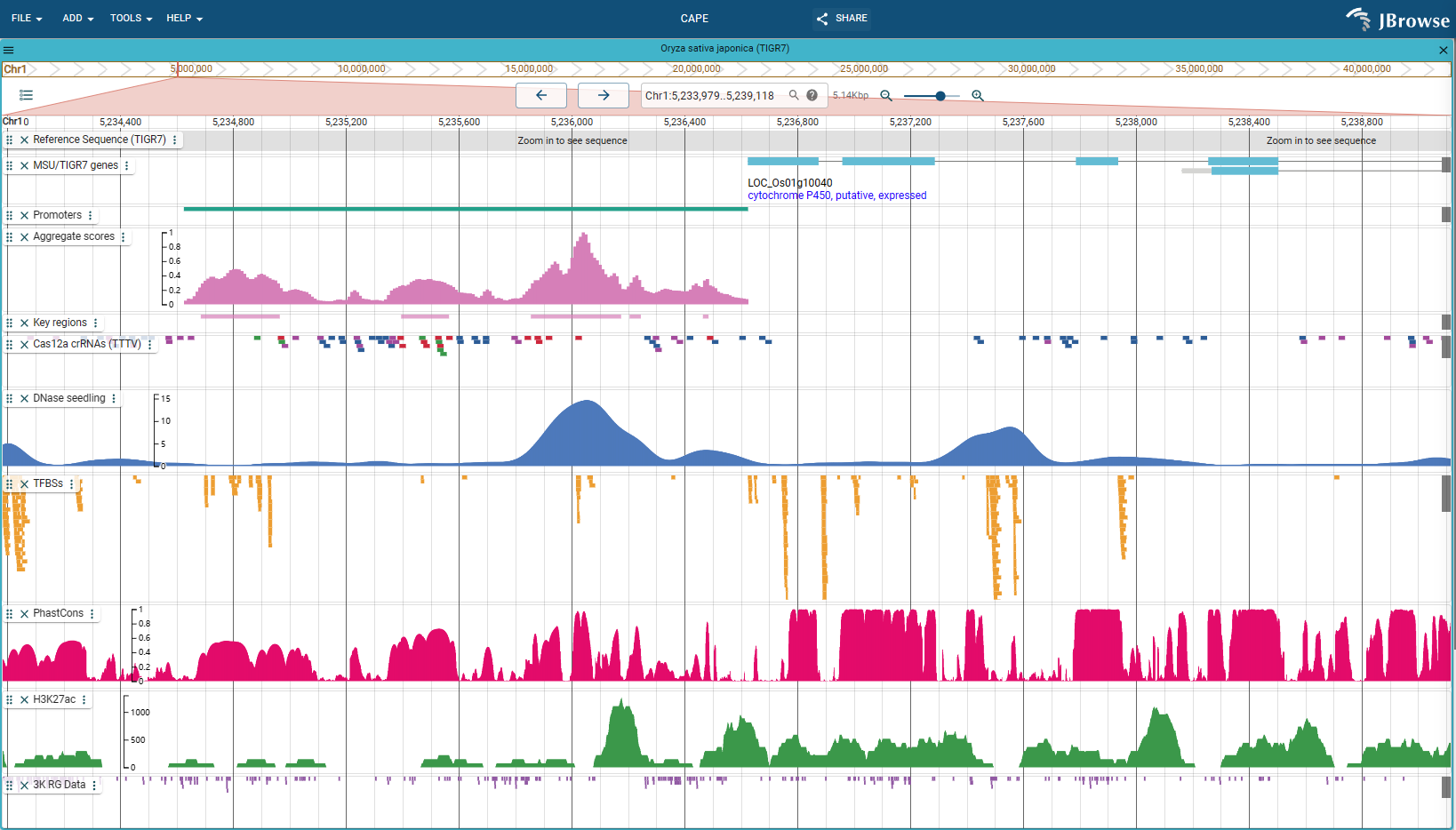
CAPE
CAPE: An efficient CRISPR-Cas12a promoter editing system for crop improvement [JBrowse preview](https://bioinfor.yzu.edu.cn/jbrowse2/cape) ## Introduction we demonstrate a **CAPE** based promoter editing and tuning pipeline for efficient production of useful quantitative...

CAPE
CAPE: An efficient CRISPR-Cas12a promoter editing system for crop improvement JBrowse preview Introduction we demonstrate a CAPE based promoter editing and tuning pipeline for efficient production of useful quantitative trait...
Lab meeting
Genome-wide cis-decoding for expression design in tomato using cistrome data and explainable deep learning
Abstract: In the evolutionary history of plants, variation in cis-regulatory elements (CREs) resulting in diversification of gene expression has played a central role in driving the evolution of lineage-specific traits....

OsAAP6 functions as an important regulator of grain protein content and nutritional quality in rice
Abstract: Grains from cereals contribute an important source of protein to human food, and grain protein content (GPC) is an important determinant of nutritional quality in cereals. Here we show...